Authors: Kexin Huang, Jaan Altosaar, and Rajesh Ranganath
Year: 2019
Original Abstract: Clinical notes contain information about patients beyond structured data such as lab values or medications. However, clinical notes have been underused relative to structured data, because notes are high-dimensional and sparse. We aim to develop and evaluate a continuous representation of clinical notes. Given this representation, our goal is to predict 30-day hospital readmission at various timepoints of admission, including early stages and at discharge. We apply bidirectional encoder representations from transformers (BERT) to clinical text. Publicly-released BERT parameters are trained on standard corpora such as Wikipedia and BookCorpus, which differ from clinical text. We therefore pre-train BERT using clinical notes and fine-tune the network for the task of predicting hospital readmission. This defines ClinicalBERT. ClinicalBERT uncovers high-quality relationships between medical concepts, as judged by physicians. ClinicalBERT outperforms various baselines on 30-day hospital readmission prediction using both discharge summaries and the first few days of notes in the intensive care unit on various clinically-motivated metrics. The attention weights of ClinicalBERT can also be used to interpret predictions. To facilitate research, we open-source model parameters, and scripts for training and evaluation. ClinicalBERT is a flexible framework to represent clinical notes. It improves on previous clinical text processing methods and with little engineering can be adapted to other clinical predictive tasks.
Research question: How can we apply BERT to clinical notes and fine-tune it for 30-day hospital readmission prediction?
Reasons for the study: Unstructured, high-dimensional, and sparse data, such as clinical notes, are difficult to use in traditional machine learning methods. Yet clinical notes contain a significant clinical value. In order to assess a patient, clinicians may need to read clinical notes to make decisions under time constraints. Therefore, processing clinical notes and making predictions from them can help in medical environments.
Data: The Medical Information Mart for Intensive Care III (MIMIC-III) dataset was used. MIMIC-III consists of the electronic health records of 58,976 unique hospital admissions from 38,597 patients in the intensive care unit of the Beth Israel Deaconess Medical Center between 2001 and 2012. There are 2,083,180 de-identified notes associated with the admissions.
Methods: ClinicalBERT is an application of the BERT model to clinical corpora to address the challenges of clinical text. Representations are learned using medical notes and further processed for clinical tasks.
BERT Model: BERT is a deep neural network that uses the transformer encoder architecture to learn embeddings for text. The transformer encoder architecture is based on a self-attention mechanism.
Data representation: A clinical note input to ClinicalBERT is represented as a collection of tokens. These tokens are subword units extracted from text in a preprocessing step. A token in a clinical note is represented as a sum of the token embedding, a learned segment embedding, and a positional embedding.
Pre-training ClinicalBERT: BERT is trained on BooksCorpus and Wikipedia. As the clinical text is different from text from books and Wikipedia, ClinicalBERT is pre-trained in text from clinical notes using as an initial checkpoint a BERT model. The pre-training process used the same tasks like the ones used in BERT.
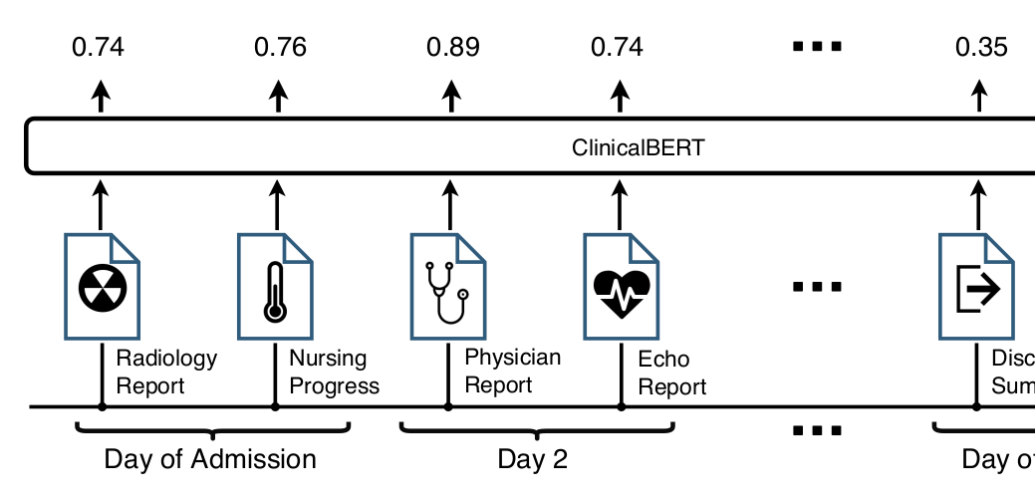
Fine-tuning ClinicalBERT: After pre-training, ClinicalBERT is fine-tuned on a clinical task: readmission prediction in the next 30 days. Given clinical notes as input, the output of ClinicalBERT is used to predict the probability of readmission. The model parameters are fine-tuned to maximize the log-likelihood of a binary classifier (if the patient will be readmitted or not). Figur
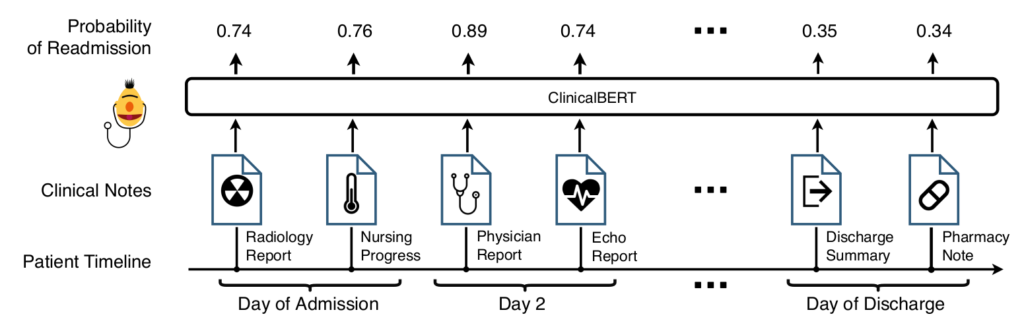
Results: There are two main lines of results, language modeling and readmission prediction. First, BERT and ClinicalBert are compared in language modeling and next sentence prediction tasks using clinical notes from MIMIC-III. The following table shows the results:

Second, for the readmission prediction task, ClinicalBert is compared to BERT and two other baseline methods, Bag-of-words and Bidirectional long short-term memory (BI-LSTM) with Word2Vec embeddings.
Bag-of-words: this method uses word counts to represent a note. The 5,000 most frequent words are used as features. Logistic regression with L2 regularization is used to predict readmission.
Bidirectional long short-term memory (BI-LSTM) and Word2Vec: a BI-LSTM is used to model words in a sequence. The final hidden layer is used to predict readmission.
The evaluation metrics are Area under the receiver operating characteristic curve (AUROC), Area under the precision-recall curve (AUPRC), and Recall at precision of 80% (RP80). The following table shows the results:
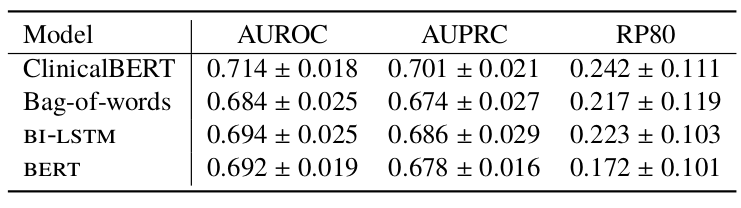
Additionally, different cutoff times were tested for readmission predictions as it is not always possible to use discharge summaries at the end of a patient’s stay. The following table shows results for models tested given notes up to 24-48h or 48-72h of a patient’s admission.
Discussion: Empirically, ClinicalBERT is an accurate language model and captures physician-assessed semantic relationships in clinical text. In a 30-day hospital readmission prediction task, ClinicalBERT outperforms a deep language model and yields a large relative increase on recall at a fixed rate of false alarms. Finally, note that the MIMIC-II dataset is small compared to the large volume of clinical notes available internally at hospitals. Rather than using pre-trained MIMIC-II ClinicalBERT embeddings, this suggests that the use of ClinicalBERT in hospitals should entail re-training the model on this larger collection of notes for better performance.