Authors: Alvin Rajkomar, Eyal Oren, Kai Chen, Andrew M. Dai, Nissan Hajaj, Michaela Hardt, Peter J. Liu, Xiaobing Liu, Jake Marcus, Mimi Sun, Patrik Sundberg, Hector Yee, Kun Zhang, Yi Zhang, Gerardo Flores, Gavin E. Duggan, Jamie Irvine, Quoc Le, Kurt Litsch, Alexander Mossin, Justin Tansuwan, De Wang, James Wexler, Jimbo Wilson, Dana Ludwig, Samuel L. Volchenboum,
Katherine Chou, Michael Pearson, Srinivasan Madabushi, Nigam H. Shah, Atul J. Butte, Michael D. Howell, Claire Cui, Greg S. Corrado, and Jeffrey Dean.
Year: 2018
Original Abstract: Predictive modeling with electronic health record (EHR) data is anticipated to drive personalized medicine and improve healthcare quality. Constructing predictive statistical models typically requires extraction of curated predictor variables from normalized EHR data, a labor-intensive process that discards the vast majority of information in each patient’s record. We propose a representation of patients’ entire raw EHR records based on the Fast Healthcare Interoperability Resources (FHIR) format. We demonstrate that deep learning methods using this representation are capable of accurately predicting multiple medical events from multiple centers without site-specific data harmonization. We validated our approach using de-identified EHR data from two US academic medical centers with 216,221 adult patients hospitalized for at least 24 h. In the sequential format we propose, this volume of EHR data unrolled into a total of 46,864,534,945 data points, including clinical notes. Deep learning models achieved high accuracy for tasks such as predicting: in-hospital mortality (area under the receiver operator curve [AUROC] across sites 0.93–0.94), 30-day unplanned readmission (AUROC 0.75–0.76), prolonged length of stay AUROC 0.85–0.86), and all of a patient’s final discharge diagnoses (frequency-weighted AUROC 0.90). These models outperformed traditional, clinically-used predictive models in all cases. We believe that this approach can be used to create accurate and scalable predictions for a variety of clinical scenarios. In a case study of a particular prediction, we demonstrate that neural networks can be used to identify relevant information from the patient’s chart.
Research question: How can we build deep learning predictive models using patients’ entire raw EHR data based on the Fast Healthcare Interoperability Resources (FHIR) format?
Reasons for the study: Deep learning can help us to build predictive models using raw EHR, and avoid the labor-intensive process of feature engineering, typically done when building traditional statistical models. This study shows that such models can predict in-hospital mortality, 30-day unplanned readmission, prolonged length of stay, and all of a patient’s final discharge diagnoses, with state-of-the-art performance.
Data: EHR data from the University of California, San Francisco (UCSF) from 2012 to 2016, and the University of Chicago Medicine (UCM) from 2009 to 2016 are included. Both datasets contained patient demographics, provider orders, diagnoses, procedures, medications,
laboratory values, vital signs, and flowsheet data, which represent all other structured data elements (e.g., nursing flowsheets), from all inpatient and outpatient encounters. The UCM dataset additionally contained de-identified, free-text medical notes.
Methods:
Data representation: A single structure was developed that could be used for all predictions. The structure represents the entire EHR in temporal order, where every patient has a timeline of events in FHIR format. Each FHIR resource had multiple elements (features). For example, a medication resource would have a feature for medication name and another one for medication RxNorm code. The values of these features could be single tokens (“Aspirin”) or a sequence of tokens (for free text). For numeric values, two representation forms were used. First, concatenate name, value, and unit (e.g “Hemoglobin 12 g/dL”) and assign this to a unique token. Second, concatenate the quantile of the value (calculated for each type of feature) and assign this to a unique token. Unique tokens of features were individually mapped to a d-dimensional embedding vector with d either tuned as a hyperparameter or derived from the number of unique tokens (token had to appear at least twice in the training set for receiving its own embedding vector, for observation code and value pairs, the token had to appear at least 100 times). Less frequent tokens were hashed and mapped to a small number of out-of-vocabulary embedding vectors. The time associated with each resource relies on the difference to the time of prediction in seconds (delta-time), however is different for every model architecture.
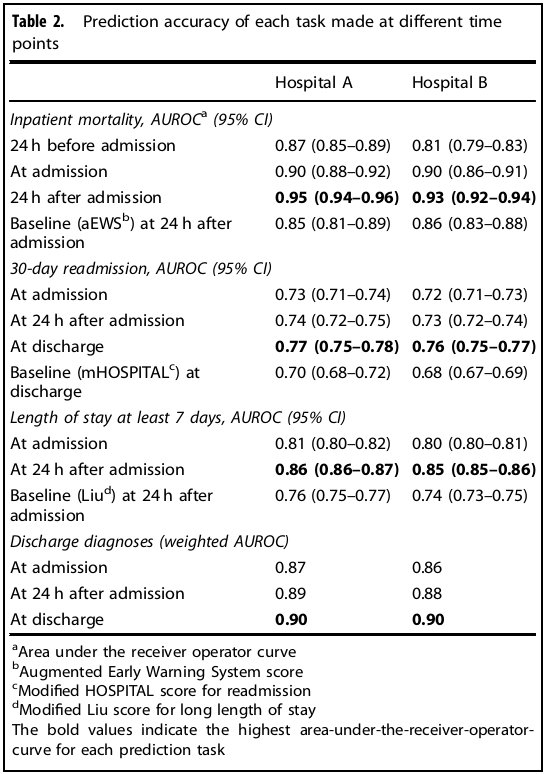
Prediction timing: For inpatient mortality, predictions were made every 12 hrs from 24 hrs before admission until 24 hrs after admission (prediction at 24 hrs after admission was comparable with other studies). Unplanned readmission and the set of diagnosis codes were predicted at admission, 24 hrs after admission, and at discharge (prediction at discharge was comparable with other studies). Long length of stay was predicted at admission and 24 hrs after admission.
Study cohort: All 18-years-old-or-older patients were included. Only hospitalizations of 24 hrs or longer were included. To simulate real-time predictions, patients typically removed in studies of readmission were included, such as those discharged against medical advice, since these exclusion criteria would not be known when making predictions earlier in the hospitalization.
Determination of unplanned readmission: The definition given by the Centers for Medicare & Medicaid Services (CMS) was used to determine planned admissions.
Main Model: Weighted Recurrent neural network model (RNN): Embeddings were used to represent features. These embeddings were weight averaged, concatenated, and an embedding for delta-time was added (see figure below).

The sequence of embeddings was reduced down by weighted averaging them in 12-hours time-steps. An LSTM network was fed with this reduced sequence. The hidden state of the final time-step of the LSTM was fed into an output layer, and the model was optimized to minimize the log-loss (logistic regression or softmax loss depending on the task). Regularization techniques such as dropout and L2 weight decay were applied. Everything was jointly optimized using Adagrad for binary tasks and Adam for multilabel tasks.
Baseline models: Every task has a baseline model. aEWS for mortality, modified HOSPITAL score for readmission, modified Liu for length of stay.
Results: Data from a total of 216,221 hospitalizations involving 114,003 unique patients were used for this study. See results in table below.
Discussion: Deep learning models for the presented tasks were more accurate than traditional models. However, the study has some limitations. First, it is a retrospective study. Second, good accuracy does not necessarily imply better health care. Finally, further research is needed to determine how well a model from one site will work in another site.
